A year or so ago, University of Florida toxicologist Chris Vulpe started getting calls, first from one colleague, then another, and another. They had an idea and wanted to use CRISPR in their labs, but had questions about this exciting new technology. Could he help?
Vulpe and his Ph.D. student Amin Sobh had come to UF from the University of California at Berkeley, one of the pioneers in CRISPR, and were familiar with the technology. As they spent more and more time assisting colleagues, it was clear Vulpe’s laboratory in the Center for Environmental and Human Toxicology could not meet the need for help with the hot new gene-editing technique.

Everyone, seemingly, wanted to use CRISPR.
“We were getting a little overwhelmed by people wanting to collaborate or asking for help. It was clear there was a surge in interest,” says Vulpe, who will trade his ad hoc CRISPR role to direct a new core starting this semester in the Interdisciplinary Center for Biotechnology Research, with dedicated staff and technical help for researchers interested in CRISPR.
It’s not that CRISPR is complicated — the opposite is true, with a little practice — but it’s innovative, the newest wave in genomics technology. Like a tsunami, it has overtaken previous methods of genetic engineering, swamping them and taking over the ecosystem of research. In 2012, PubMed, an archive of scientific publishing, listed only 127 articles on CRISPR. By 2017, 3,185 had been published.
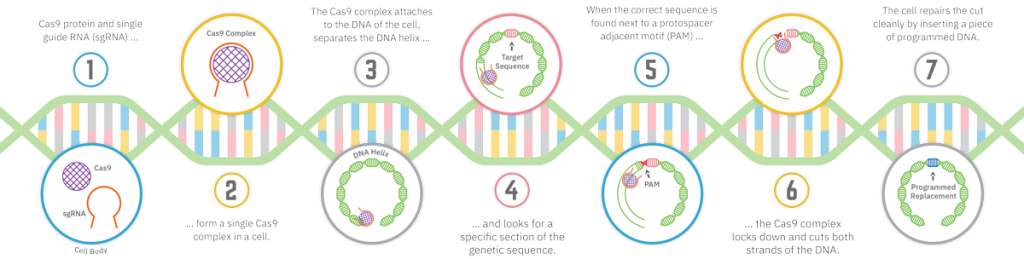
CRISPR has passed gene-editing technologies that were new themselves: zinc finger nucleases, or ZFNs, and transcription activator-like effector nucleases, or TALENs. Scientists describe CRISPR as elegant, and even its acronym, pronounced crisper — a real word — beats the clunky ZFN and TALEN. CRISPR stands for clustered regularly interspaced short palindromic repeats (see illustration), a mouthful to be sure. But it is so straightforward and inexpensive that biohackers are using it in their garages and kitchens.
“There’s a guy I follow online who does this at home,” says horticulture department Chair Kevin Folta, an expert in strawberry genetics. “He wants to make purple flowers, and he reaches out occasionally for advice.
“It’s kind of the wild west of gene editing.”
CRISPR can be breathtakingly precise. Gene editing with CRISPR is so exact, it’s like going into a book, editing one word and changing the end of the story, Folta says. Imagine removing the word “not” from the Ten Commandments. A genome is a library, too, with genes that carry instructions for each cell. With CRISPR, a single gene can be targeted at any point in the genome.
Guide RNA can be programmed to find a point, and carry along Cas9, a protein that recognizes its target and cuts both strands of DNA. Not only can genetic material be removed, but missing genetic information can be edited in. A modified Cas9 gene can be used to turn a gene on or off temporarily, enhancing or muting its expression and allowing a scientist to examine the function of the gene without editing it. Other proteins, too, have been constructed to achieve different gene-editing goals.
“CRISPR has changed significantly in the last three years. The newest methods are even better — faster, more agile, predictable, specific — that’s what makes it so powerful,” Folta says. “We’re seeing the normal expansion of science happening. This is germane to everybody who looks at a cell.”
CRISPR and Human Health
Jonathan Licht, director of the UF Health Cancer Center and a specialist in leukemia, lymphoma and multiple myeloma, is quite familiar with the pace of research with CRISPR. In the last couple of years, his lab was poised to publish work on CRISPR, only to open scientific journals and find the lab’s idea in print.
Licht began using CRISPR when a colleague at a symposium suggested he look into it, and he tasked a postdoc with researching CRISPR.
“Every week in our lab meeting, we had the CRISPR report,” says Licht, and ideas poured out.
“It has unleashed a creativity in biomedical science, cancer science, neuroscience,” Licht says. “The things you can do with CRISPR are amazing.”
One way Licht uses CRISPR is to study a gene called NSD2, which codes for a protein that activates other genes. About 10 to 15 percent of children with acute lymphocytic leukemia who relapse early in therapy have a mutation in NSD2. Licht wanted to see if he could use CRISPR/Cas9 to remove the mutant gene without affecting the remaining normal genes. Previously, he used RNA interference to do this, but that method was imprecise, inhibiting both the mutant protein and the normal protein. The mixed effect made it difficult to isolate what the mutant protein was doing.

With CRISPR, he was able to cleanly edit the gene.
Using CRISPR in three acute lymphocytic leukemia cell lines in culture, Licht’s lab was able to compare the biology of mutant cells with that of cells edited to remove the mutation. In all three lines, the activity of the edited cells quieted: they grew more slowly, were less mobile and invasive, and became more sensitive to commonly used cancer therapies.
When studied in mice, NSD2 mutant cells rapidly caused animals to become ill with a particular accumulation of cells in the brain, while the cells with the mutation removed were less aggressive and spread more evenly through the animals, taking up to a week longer to make the animals ill.
Licht found that the mutant NSD2 protein was changing leukemia cell behavior by activating a distinctly abnormal set of genes more characteristic of the nervous system than blood cells. The experiments suggest that leukemia cells that gain this mutation have a growth advantage that allows them to survive therapy and cause relapse. Licht hopes to use the new cells his group created to find new therapies to treat resistant childhood leukemia.
Licht is getting ready to collaborate with Vulpe, who has developed a “druggable genome library” of CRISPR sequences. The library allows every protein for which there is a Food and Drug Administration (FDA)-approved drug to be eliminated from the cell. Using the library, Licht says, he can determine which proteins are critical for the growth of NSD2 mutant leukemia cells, then conduct experiments using drugs that work against the proteins.
Vulpe says the beauty of that approach is that instead of targeting one gene at a time, all the genes in a genome can be investigated together, or a specific subset of genes of interest can be tested.
“What’s different is the ability to look at many genes at once,” Vulpe says.
Toxicology
Back in his toxicology lab in the College of Veterinary Medicine, Vulpe hopes the druggable genome library will lead to an explosion of much-needed knowledge on chemicals and toxicants.
“Over 100,000 chemicals are used in the world, and how many do we know anything about?” Vulpe asks. “Maybe 1,000. So, 99 percent of all chemicals being used every day that you are exposed to right now, we don’t know anything about.
“And there’s this perception that chemicals are regulated, and that perception is largely false,” Vulpe says.
While the FDA has to prove something is safe to approve it, until recent reforms began, the Environmental Protection Agency (EPA) had to prove something was unsafe before it could be banned. Only about six chemicals have ever been federally banned, Vulpe says. Hexavalent chromium, one of the most potent known carcinogens, is only banned from cooling towers on tops of buildings but can be used on playgrounds in treated wood, Vulpe says. Chemicals get manufactured, put out into the environment, and then it’s essentially a game of catchup to prove a chemical is dangerous.
“I tell my students, we’re undergoing the giant chemical experiment of the 21st century. Take Earth, pour 100,000 chemicals on it, and see what happens in 20 or 30 years,” Vulpe says.
Vulpe thinks a more scientific approach to toxicity testing is needed.
“We could test every chemical using standard protocols, but it would take hundreds of years and cost trillions of dollars,” Vulpe says. “Or we can use new approaches like CRISPR.”
To understand the basic mechanisms of toxicity, Vulpe’s lab created a library of cells, with a single different gene knocked out in each cell. For example, in a library of 100 cells, 100 different genes are knocked out. The idea is to expose each cell to a sub-lethal dose of a toxicant. The cells can then be examined to see which resist exposure to a toxicant and which succumb to exposure to a toxicant. This technique, called genome-wide screening, allows researchers to understand the function of each gene in relation to toxicity.
“There’s a burgeoning interest now in what this technology can do for understanding toxicity,” Vulpe says.
With a grant from the EPA, Vulpe is developing ToxCRISPR, a library focused on the 4,000 genes in mammals that function as toxicant pathways, whether it’s uptake, detoxification or transport.
Before CRISPR, the standard way to test toxicity was on animals. But to test every chemical on a mouse, for instance, would be impractical. CRISPR, Vulpe says, is a next step in the evolution of trying to develop in vitro approaches to understanding how chemicals work. What takes years to accomplish in mice, takes weeks in the cell libraries, and bypasses the issue of animal testing.
Vulpe says researchers still don’t know the function of about half the genes in the human genome.
“There’s that gene, it’s got to be important, but we really don’t know what it does. We can use CRISPR to make the connection between genes and their function,” Vulpe says. “We can do things we could not do before, so we start thinking about things in a completely new way.”
Moving CRISPR Forward
Vulpe met one of his CRISPR colleagues by accident. He was out running in San Felasco Hammock when he met Chris Reisch, a microbiologist recently transplanted to Gainesville from his postdoc at the Massachusetts Institute of Technology, another pioneer in CRISPR research.
Vulpe says Reisch has moved beyond Cas9 and developed completely new CRISPR tools, including one other labs worldwide have requested more than 200 times from Addgene, a nonprofit repository where researchers send plasmids — genetic cell structures — so that other researchers can use them. Reisch’s plasmid goes beyond even the precision of editing a single gene and allows a gene to be edited or changed one or two base pairs at a time.

Recently, he submitted another plasmid to Addgene, adding another level of precision to gene editing. This plasmid allows for transcriptional repression, in which a gene is targeted with a variant of Cas9 that is no longer capable of cutting the DNA. Instead, it sits on the DNA, and that prevents other proteins within the cell from binding with the DNA to express the gene.
“The gene is still physically there, it just can’t be expressed,” Reisch says. “This is a powerful tool because you don’t have to edit the chromosome. Everything that is happening is transient and you can do it quickly. You put in the CRISPR system, target the specific gene, and turn it off.”
The method is particularly valuable in studying essential genes, the genes that must be present and cannot be cut with CRISPR/Cas9 without killing the cell. With transcriptional repression, Reisch can turn off an essential gene and see what happens to the cell when that gene and its proteins are missing, then turn it back on.
“This is a way to look at those genes for which we still don’t know the function,” Reisch says. “We can knock down the essential, but unknown, genes and see what they’re actually doing.”
Before CRISPR, Reisch says, it might have taken a few months to do a gene deletion, so a researcher might hesitate before embarking on that process. What if he or she deleted a gene thinking it had one function and then found it had a different function? Depending on the study at hand, the researcher might have to start over. CRISPR removes that hesitation.
“We were slogging before CRISPR,” Reisch says. “In grad school, it would take months to make a single gene deletion. Now, we could conceivably do 10 or 20 gene deletions in a week.”
CRISPR removed some of the limitations of lab work, Reisch says, but created others.
“We have the ability to do a lot of work in the lab, but now analyzing the data, doing the actual science, takes months.”
A CRISPR Rescue
When CRISPR got hot a few years ago, researcher Nian Wang at UF’s Citrus Research and Education Center immediately thought of using it to battle two threats to the Florida citrus industry: canker and greening.
He had been using the elaborate and labor-intensive TALENs, and switched to cheaper and simpler CRISPR methods to speed up his work. In proof-of-concept studies, he has already shown that gene editing with CRISPR can create a variety of grapefruit that is resistant to canker. Afterward, the Florida Legislature awarded Wang $1 million to deploy CRISPR in citrus greening, and the USDA recently followed suit with a $3.6 million award. Citrus greening, also known as Huanglongbing, is a scourge that growers fear could be the end of the $9 billion-a-year citrus industry in Florida.

But Wang is hopeful. He anticipates success on two fronts: generating greening-resistant varieties and greening-tolerant varieties, meaning the trees might have greening but would tolerate the infection and yield healthy fruit.
The other advantage of CRISPR, Wang says, is that it uses an organism’s own genome, so avoids controversies over transgenic technologies, such as putting spinach genes in a citrus tree, or animal genes into a plant. Since CRISPR turns off the gene that makes citrus susceptible to greening, rather than borrowing a gene from spinach, it would not face the regulation of a genetically modified organism, or GMO, and could be commercialized immediately. That would be welcome relief for citrus growers.
Florida produced 242 million boxes of oranges in the 2003-2004 season. That dropped to 81.6 million boxes in 2015-16.
For Folta, the horticulture department chairman, time is of the essence.
“To breed an orange takes many years and many generations. If people in 2050 want to taste an orange, well, there are six generations of orange trees we can work with between now and then,” Folta says. “But to make the gene edits that can allow oranges to survive the current threats might take two years.
“So CRISPR allows scientists to respond to an existential threat like greening with an agility and speed we never had before.”
In his work on strawberry genetics, Folta says his lab members are dealing with the realization that experiments they started last year might have to be redone if they want to maximize the speed of CRISPR. Something that took six months last year takes two weeks now.
“I spent a summer of my graduate career sequencing 10 kilobases of DNA, 10,000 bases in each direction,” Folta says. “Now that can be done overnight for $30. The question is, now that the wheel has been reinvented, do you go back and use the reinvented wheel?”
Sources:
- Christopher Vulpe, Professor of Physiological Sciences
- Jonathan Licht, UF Health Cancer Center Director and Marshall E. Rinker Sr. Foundation and David B. and Leighan R. Rinker Chair
- Kevin Folta, Professor and Chairman, Department of Horticultural Sciences
- Christopher Reisch, Assistant Professor of Microbiology and Cell Science
- Nian Wang, Associate Professor of Microbiology and Cell Science
This article was originally featured in the Spring 2018 issue of Explore Magazine.

