A prospective cohort study by PRISMAp
Breaking barriers: PRISMAp researchers, in collaboration with several departments at the University of Florida including Biomedical Engineering, Surgery, UFHealth, and i-Heal, have just completed the first ever study that uses urinary cellular gene expression to study sepsis.

Kidneys are Key
Kidneys clear toxins from both internal and external products in the body, filtering our blood every day, every minute. They also produce hormones and serve as a control system for acid, salt, and several physiological processes. Kidney dysfunction can cause other organs to dysfunction, therefore it is key to advance research of kidney processes. Since 1 out of 2 patients with sepsis develop kidney dysfunction, early detection of sepsis can provide opportunities to minimize its life-threatening impacts.
Utilizing Urine
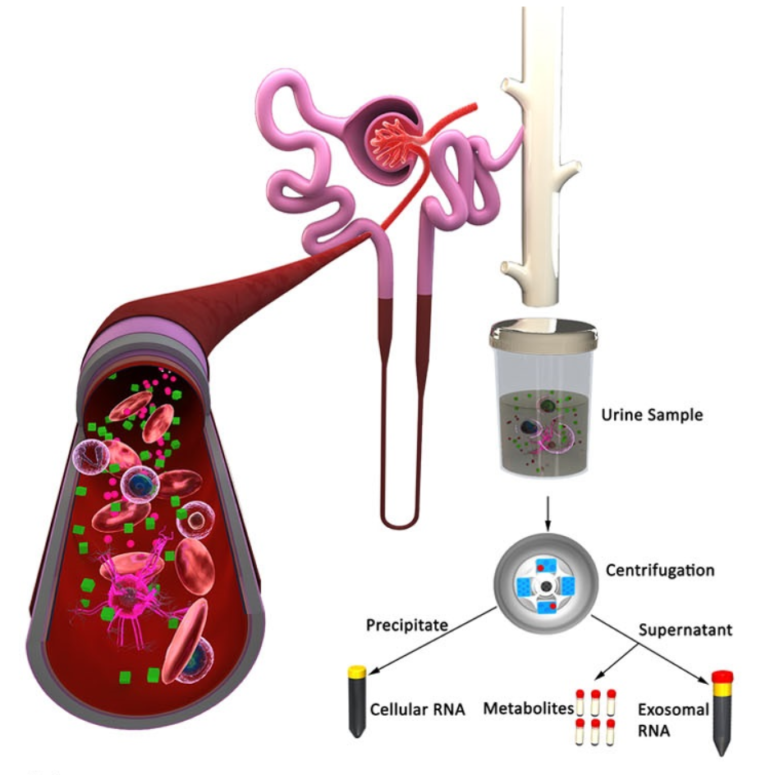
For our research study, urine was collected within 12 hours of sepsis onset to study the early sepsis complex. Why urine?
Urine is a readily available bio-fluid in the clinical setting which is rich in cells and nucleic acid. The cells in urine include both kidney cells and immune (blood) cells, so urine allows us to see what’s going on in the blood.
The goal of this study was to find out if changes in urinary cells can reflect systemic immune and kidney-specific processes that are typically only discovered using transcriptomic analyses of blood samples.
Artificial Intelligence
Artificial intelligence tools are helping us understand sepsis on a human-based level. Previous sepsis research conducted on animals did not translate well to human beings, and a lot of therapies that researchers have tried have failed. Our research is dedicated to using human samples and applying Artificial Intelligence approaches to create comprehensive data sets.
We used an ensemble of machine learning algorithms to find the subsets of genes that identifies sepsis patients from controls. We applied Machine Learning and Text Mining to whole-genome transcriptomic profiles of messenger RNA isolated from urinary cells to find genes that mark early stages of sepsis, and used Functional Analyses to index their associated pathways.
Results
We used a pathway overlap graph which connects pathways having ten or more genes in common to show that early sepsis is distinguished by five major pathway clusters associated with innate immunity, cell cycle, cell morphology and motility, hypoxia, & production of reactive oxygen species.

Analyses of cell proportions using in silico deconvolution showed significant increases in neutrophils and monocytes, as well as epithelial cells from all nephron regions in sepsis patients.
Our machine learning algorithms were trained on the discovery cohort to produce three feature lists consisting of 233, 64 and 42 probes most capable of classifying sepsis patients from controls. Performance was demonstrated on an independent external validation cohort.

Conclusions and Implications
Our study provides a new window into the pathophysiology of sepsis and marks a pioneering step towards the identification of biomarkers of sepsis from urine. This technology of using RNA from urine cells to study an entire genome was previously used only for transplantation. Despite its advantages, urine has been under-utilized in diagnosis and clinical decisions.
We hope that the momentum of this study will open the door to new possibilities using urine to detect early sepsis. Our long-term aspiration for the future is to improve accessibility of patient care by having simple bedside tests at home to determine whether they will need more medical attention. Improving offsite, at-home health monitoring provides a better quality of life for patients while also reducing the burden of medical care by detecting early signs of critical illness.
How the Magic Happens
Dr. Azra Bihorac, lab director of PRISMAp, describes these developments of new techniques using artificial intelligence to improve health care as a “multidimensional biohack.” The secret to success is expanding cross-disciplinary research to propel us towards innovation.
PRISMAp collaborates with specialists from several different departments and teams including surgery, critical care, nephrology, molecular biology, biomedical engineering, artificial intelligence, and more. This cross-disciplinary partnership is only possible because we have a diversity of dedicated professionals all in one place at the University of Florida.
When it comes to medical research, teamwork really does make the dream work.

This story originally appeared on UF PRISMAp, College of Medicine.
Check out other stories on the UF AI Initiative.